Visualization of Protein Structures
Step into the captivating world of protein structures, where scientific exploration meets artistic expression. Through the visualization tools like Pymol, Swiss PDB viewer, and the ray tracing prowess of POVRay, here is a gallery of the protein structures that I have created during my research career.
Calmodulin: A Dance with Calcium
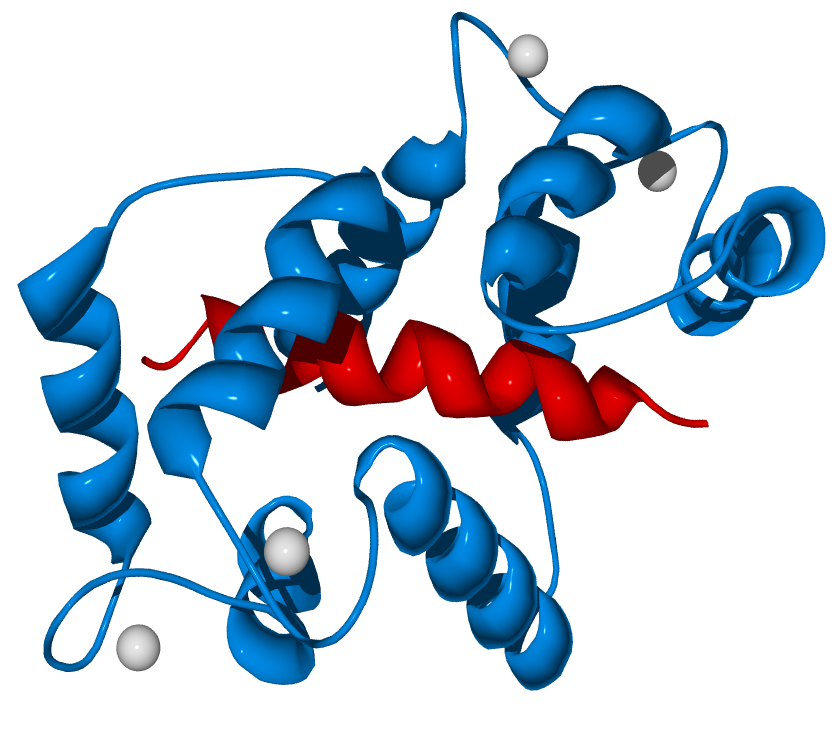
Calmodulin Binding to Calcium: Calmodulin is a protein which could binds to four calcium ions, rendered as gray spheres.
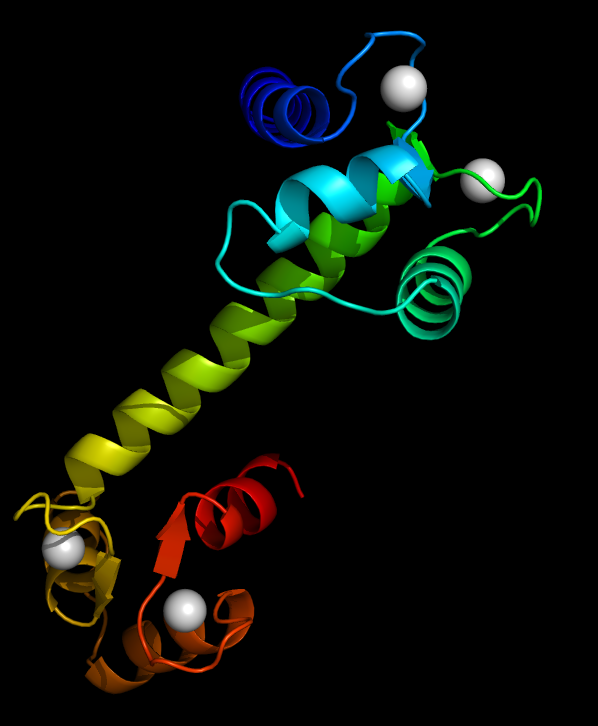
Calmodulin Binding to Ryanodine Receptor: Calmodulin can bind to an array of proteins and regulates their funcitons.
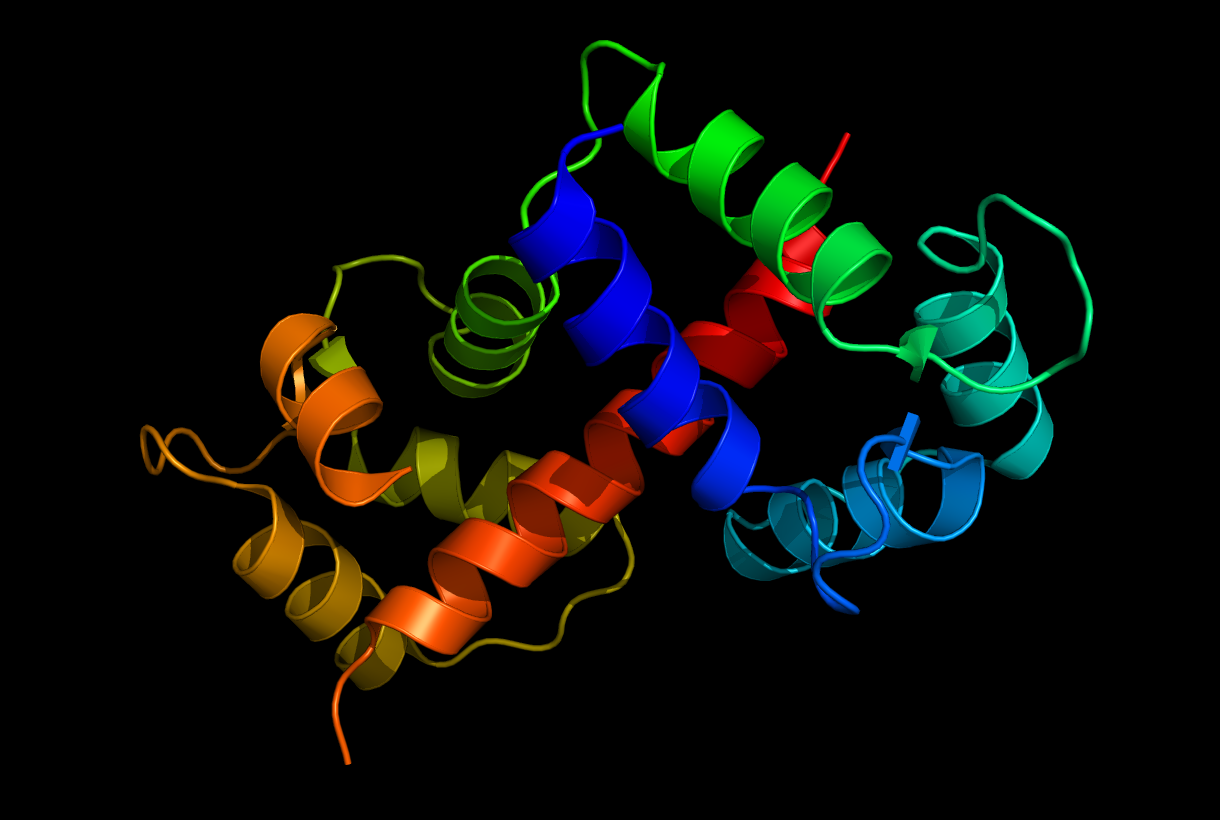
Calmodulin Binding to M13: The M13 peptide is a well-studied target of Calmodulin.
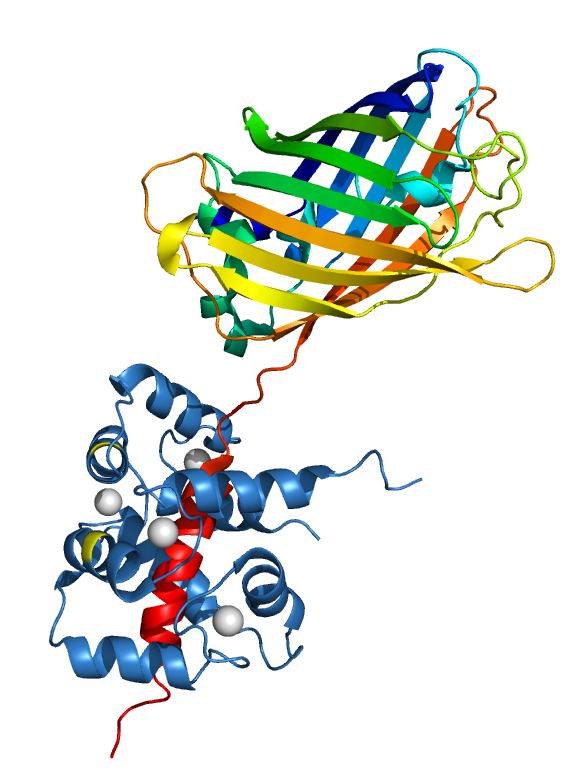
EGFP Fused with M13 Binding to Calmodulin: We have created a fusion protein of GFP and M13. Please notice this structrue was an illustrative cartoon of the structure created with Pymol.
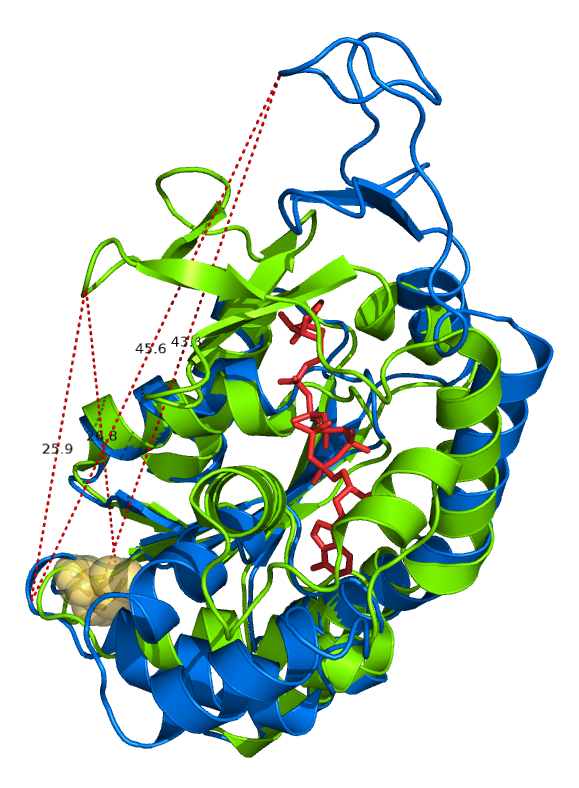
Overlaped structures of Adenylate Kinase with/without binding to the substrate analog AP5’A
The red stick model represents the substrate analog AP5’A. The visualization also incluldes a few measurements of the distance between residuals VAL148 to GLU 75 or CYS77. CYS 77 was rendered as yellow spheres.
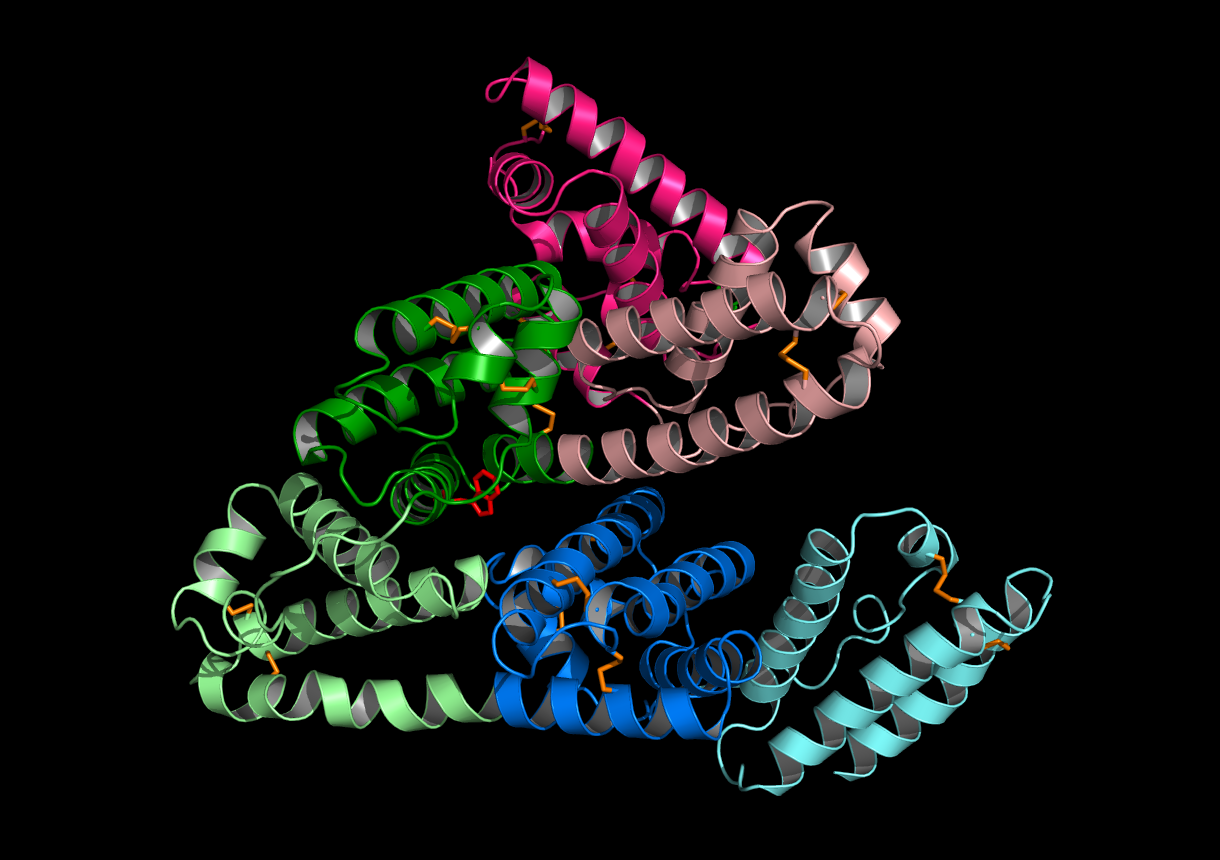
Human Serum Albumin
Human Serum Albumin (HSA) structure with all disulfide bonds highlighted as orange sticks. The tryptophan was also highlighted as red sticks. HSA is an all alpha helix protein. The alpha helice were colored by HSA’s subdomains(IA,IB,IIA,IIB,IIIA and IIIB).
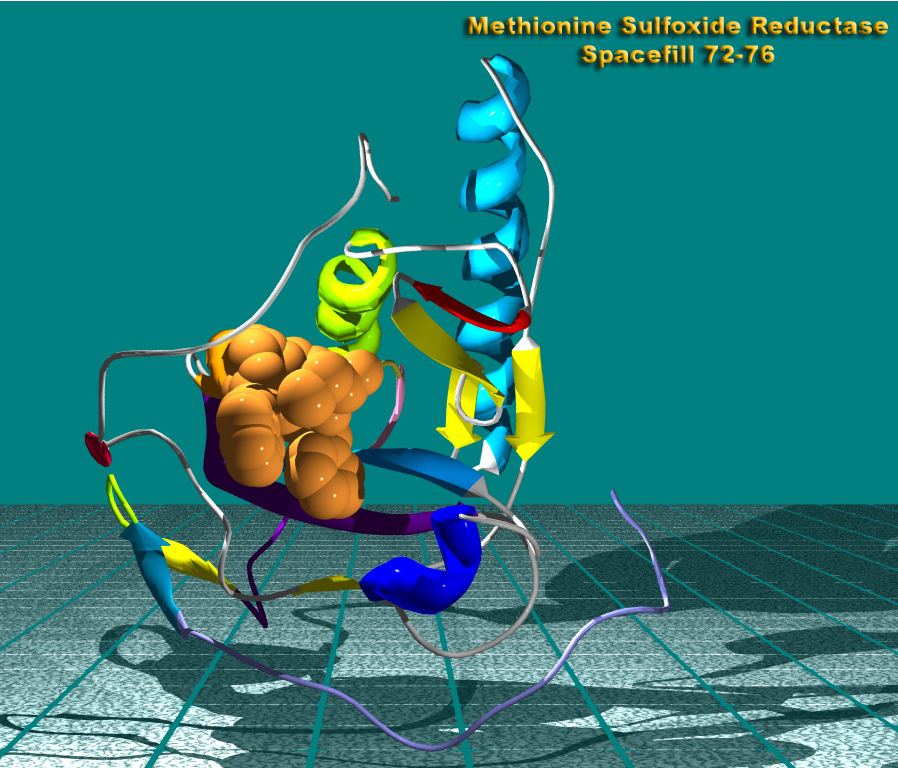
Methionine Sulfoxide Reductase A
Structure of the Methionine Sulfoxide Reductase A with enzyme activity center rendered as spheres.
Calcium ATPase
The structure of Calcium ATPase with all Tyrosines rendered as spheres. This picture was also rendered with POVray with background and clouds added.
